 |
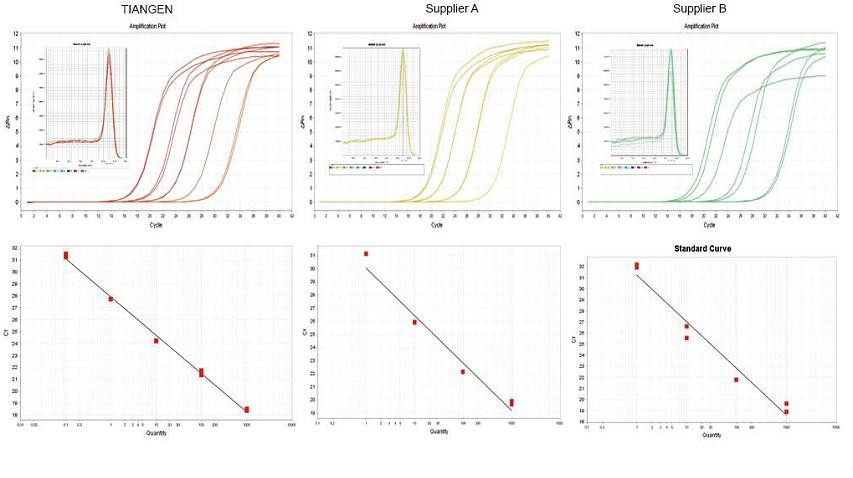
Experimental example 1. cDNA was synthesized using one-step reverse quantitative reagents of TIANGEN FastKing gDNA Dispelling RT SuperMix, relevant products from Supplier A and Supplier B respectively. Detect the RN5 gene of mice using TIANGEN Talent qPCR PreMix (SYBR Green), and the amplification curve, melting curve and standard curve were analyzed. The results show that TIANGEN FastKing gDNA Dispelling RT SuperMix has the highest quantitative Ct value after reverse transcription and excellent stress resistance, and has obvious advantages for templates with high impurity residues. |
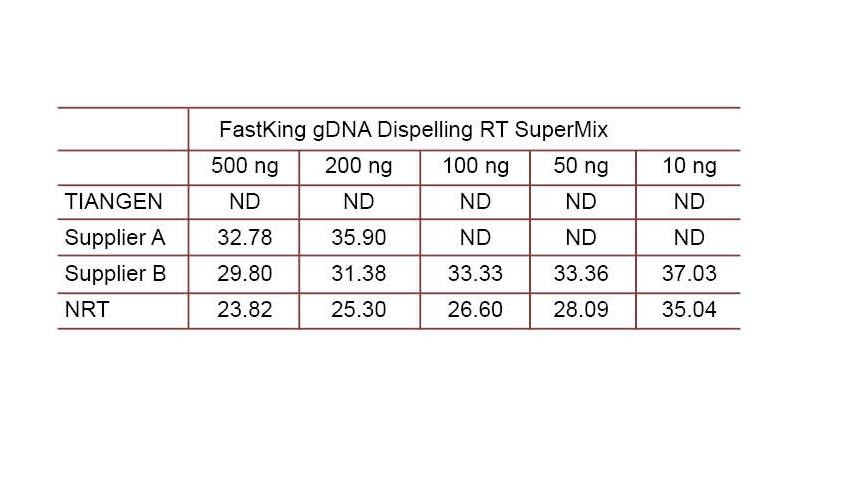 |
Experimental example 2. cDNA was synthesized using one-step reverse quantitative reagents from TIANGEN FastKing gDNA Dispelling RT SuperMix, relevant products from Supplier A and Supplier B, respectively. Detect human HsG gene using TIANGEN Talent qPCR PreMix (SYBR Green), and manually add different concentrations of genomic DNA to detect the gDNA removal ability of different reagents. Ct results show that TIANGEN FastKing gDNA Dispelling RT SuperMix has excellent ability to remove genomic DNA. Up to 500 ng of genomic DNA residue can be removed perfectly without affecting the results. ND: Not detected. NRT: Detection of the mix without reverse transcription. |
FastKing gDNA Dispelling RT SuperMix
Features
■ Fast: One-step to complete genome removal and efficient reverse transcription within 18 minutes by only adding templates.
■ High efficiency: The reverse transcriptase is modified with a hydrophobic motif, with RT efficiency exceeding 95%.
■ Simple and easy: The exclusive thermosensitive DNase has fast effect, high efficiency with shorter reaction time, and will not affect cDNA.
Specification
Type: Gene modified reverse transcriptase, gDNase
Procedures: One-step (genomic DNA removal and RT)
RT efficiency: >95%
Template: 1 ng- 2 μg
Operation time: ~18 min
Applications: The reverse transcribed cDNA can be used in conventional PCR, Real time PCR, cDNA library construction, SAGE (Serial Analysis of Gene Expression), primer extension and other conventional experiments.
All the products can be customized for ODM/OEM. For details, please click Customized Service(ODM/OEM)
Experimental Example
FAQ
A-1 RNA is degraded
——Purify high quality RNA with no contamination. The material from which RNA is extracted should be as fresh as possible to prevent RNA degradation. Analyze RNA integrity on denatured gel before RT reaction. After RNA extraction, it should be stored in 100% formamide. If RNase inhibitor is used, the heating temperature should be <45°C, and the pH should be less than 8.0, otherwise the inhibitor will release all bound RNase. Moreover, RNase inhibitor should be added in solutions containing ≥ 0.8 mM DTT.
A-2 RNA contains inhibitors of reverse transcription reactions
——Reverse transcription inhibitors include SDS, EDTA, glycerol, sodium pyrophosphate, spermidine, formamide, guanidine salt, etc. Mix the control RNA with the sample, and compare the yield with the control RNA reaction to check whether there is an inhibitor. Wash RNA precipitation with 70% (v/v) ethanol to remove inhibitors.
A-3 Insufficient annealing of primers used for synthesizing the first strand of cDNA
——Determine that the annealing temperature is suitable for the primers used in the experiment. For random hexamers, it is recommended to keep the temperature at 25°C for 10 min before reaching the reaction temperature. For gene-specific primers (GSP), try other GSP, or switch to oligo(dT) or random hexamer.
A-4 Small amount of starting RNA
——Increase the amount of RNA. For RNA samples less than 50 ng, 0.1 μg to 0.5 μg acetyl BSA can be used in the first strand cDNA synthesis
A-5 The target sequence is not expressed in the analyzed tissues.
——Try other tissues.
A-6 PCR reaction fails
——For two-step RT-PCR, the cDNA template in the PCR step cannot exceed 1/5 of the reaction volume.
A-1 Non-specific annealing of primers and templates
——The 3’-end of primers should not contain 2-3 dG or dC. Use Gene-specific primers in the first strand synthesis instead of random primers or oligo(dT). Use a higher annealing temperature in the first few cycles, and then a lower annealing temperature. Use hot-start Taq DNA polymerase for PCR to improve the specificity of the reaction.
A-2 Poor design of gene-specific primers
——Follow the same principles for amplification primer design.
A-3 RNA contaminated with genomic DNA
——Treat RNA with PCR-grade DNase I. Set up a control reaction without reverse transcription to detect DNA contamination.
A-4 Forming of primer dimer
——Design primers without complementary sequences at the 3’ end.
A-5 Too high Mg2+ concentration
——Optimize Mg2+ concentration for each template and primer combination
A-6 Contaminated with foreign DNA
——Use aerosol-resistant tips and UDG enzymes.
A-1 The content of the first strand product is too high
——Reduce the amount of the first strand product in the conventional PCR reaction step.
A-2 Too high primer amount in PCR reaction
——Reduce primer input.
A-3 Too many cycles
——Optimize PCR reaction conditions and reduce PCR cycle number.
A-4 Too low annealing temperature
——Increase annealing temperature to prevent non-specific initiation and extension.
A-5 Non-specific amplification of oligonucleotide fragments generated by DNase degradation of DNA ——Extract high-quality RNA to prevent DNA contamination.
RT-PCR is to reverse transcribe RNA into cDNA, and then use the reverse transcribed cDNA as a template for PCR reaction to amplify the target fragment. Choose either random primers, Oligo dT and gene specific primers according to the specific conditions of the experiment. All the above primers can be used for short eukaryotic cell mRNA without hairpin structure.
Random primer: Suitable for long RNA with hairpin structure, as well as all kinds of RNA such as rRNA, mRNA, tRNA, etc. They are mainly used for RT-PCR reaction of single template.
Oligo dT: Suitable for RNA with PolyA tailing (prokaryotic RNA, eukaryotic Oligo dT rRNA and tRNA do not have PolyA tails). Because Oligo dT is bound to PolyA tail, the quality of RNA samples is required to be high, and even a small amount of degradation will greatly reduce the amount of full-length cDNA synthesis.
Gene-specific primer: Complementary to the template sequence, suitable for situations where the target sequence is known.
There are two ways:
1. Internal reference method: In theory, cDNA is DNA fragments of different lengths, so the result of electrophoresis is smear. If RNA abundance is low, no product will show in electrophoresis, but this does not mean no product will be amplified by PCR. In general, internal reference can be used to detect cDNA. If the internal reference has results, the quality of cDNA can be basically guaranteed (in a few cases, if the target gene fragment is too long, there may be exceptions).
2. If there is a known gene amplified by this template, it can be verified by the primers of this gene. The amplification of internal reference does not necessarily mean that there is no problem with cDNA. Because internal reference has high abundance in cDNA, it is easy to amplify. If cDNA is partially degraded for various reasons, from the perspective of probability, PCR results of low abundance target genes will be greatly affected. While internal reference is still high in abundance, the amplification will probably not be affected.
Partially degrade of RNA. Detect the integrity and purify of RNA
The RNA contents of different species may be different, but in general, the extracted total RNA should contain two clear 28S and 18S bands in gel electrophoresis, and the brightness of the former band should be twice as high as that of the latter. The 5S band indicates that RNA has been degraded, and its brightness is proportional to the degree of degradation. The successful amplification of internal reference does not mean that there is no problem with RNA, because the internal reference is in high abundance, RNA can be amplified so long as the degradation is not severe. The OD260/OD280 ratio of pure RNA measured by spectrophotometer should be between 1.9 and 2.1. A small amount of protein impurity in RNA will reduce the ratio. As long as the value is not too low, RT will not be affected. What matters the most for RT is RNA integrity.
The extension of the internal reference gene can only indicate that RT has succeeded, but it is not necessarily related to the quality of the cDNA strand. Because the internal reference fragments are generally small in size and high in expression, they are easier to be successful in reverse transcription. However, the size and expression of target gene varies from gene to gene. The cDNA quality cannot be judged only by internal reference especially for the target fragments longer than 2 kb.
Some samples have complex secondary structures, or have rich GC content, or are precious with low abundance. In these cases, appropriate reverse transcriptase should be selected according to the size of the target fragment and the sample. For RNA templates with high GC content and complex secondary structure, it is difficult to open the secondary structure at low temperature, or with common reverse transcriptase . For these templates, Quant Reverse Transcriptase can be selected, since its reverse transcription performance is obviously better than that of M-MLV series reverse transcriptase, which can reverse transcribe various RNA templates efficiently and transcribe RNA into cDNA first strand to the maximum extent. When using general reverse transcriptase kit, 20 μl system can only effectively reverse transcribe 1 μg of total RNA. Please pay attention to the maximum RT capacity of kit. If the template is added in excess, reverse transcription will favor the RNA with high abundance. Therefore, it is better not to exceed the maximum capacity of the system.
A-1 Determine if RNA is degraded severely and if RT is successful
In general, the reason for the failure of internal reference amplification is often caused by serious RNA degradation. Another possible reason is reverse transcription failure. Internal reference cannot be used as a standard to judge the quality of cDNA single strand, but it can be used as a standard to judge whether reverse transcription is successful if there’s no problem of the RNA quality. The most important thing in the reverse transcription process is to maintain a constant temperature and a constant reaction system in order to improve the reaction efficiency.
A-2 Determine whether the primers for amplifying internal reference genes are reliable and if there’s any problems with reagents used in PCR.
For relative quantification, RNA must be quantified before reverse transcription, which is also required in many reverse transcription kits, for example, quantify the RNA input as 1 μg. Since the reverse transcribed cDNA is a mixed solution, including RNA, oligo dT, enzyme, dNTP, and even a little DNA residue, deviation will be caused, so it is impossible to accurately quantify the cDNA. Therefore, RNA quantification is necessary. Considering the reverse transcription efficiency is the same among different samples, the amount of cDNA obtained should be the same, and the quantitative analysis can show the comparison of expression levels of different genes in the same amount of total RNA. When performing relative fluorescence quantitative PCR, quantitative cDNA may not be required after reverse transcription because the internal reference gene can be acted as reference.
It is mainly related to the genes, and reverse transcription of long fragment is not feasible for most genes. Firstly, the efficiency of reverse transcription is far lower than that of PCR. Secondly, the GC rich region and secondary structure of many genes restrict both reverse transcription and PCR. Finally, the fidelity and amplification efficiency of PCR are difficult to guarantee at the same time. In the process of reverse transcription, no one can guarantee to get long fragment for low copy genes, especially using oligo dT. As for 5’ UTR with more GC, it is even more difficult. Therefore, it is still a reasonable method to reverse transcript with random primers, find the natural cleavage sites in the target fragment, amplify by segments, and then perform the restriction digestion and ligation. In general, it is difficult to directly amplify fragments larger than 2 kb, but it is not always impossible to obtain: 1.First of all, guarantee the integrity of RNA/mRNA, and TRIZOL extraction is preferred. 2.M-MLV RT-PCR kit can be directly used. Extend annealing time and increase cycle number in the amplification process properly. Alternatively, nested PCR can be applied, or carry out one or two reactions first with appropriately extended denaturation and extension time before normal PCR amplification, which may help to extend fragments. Pay attention to the fidelity of the polymerase. 3.Long Taq can be used in PCR to obtain ideal results. 4.For protein expression application, high fidelity polymerase should be applied.
There are two kinds of reverse transcriptase offered by TIANGEN: Quant/King RTase and TIANScript M-MLV. The main difference between them is input amount of templates. Quant is a unique reverse transcriptase, which is different from the commonly used M-MLV derived from Moloney murine leukemia virus. Quant is a new high-efficiency reverse transcriptase recombinantly expressed by engineering Escherichia coli. Quant is suitable for amplifying 50 ng-2 μg of RNA with high reverse transcriptional activity and high yield. Compared with ordinary MMLV or AMV, Quant’s biggest characteristic is that it has very strong affinity with RNA templates and can reverse transcript complex templates without high temperature denaturation. For templates with higher GC content, the reverse efficiency is higher. However, this reverse transcriptase has RNase H activity, which may affect the length of cDNA product (suitable for < 4.5 kb templates). For conventional reverse transcription, TIANScript MMLV reverse transcriptase is recommended. This RTase is a modified enzyme with very weak RNase H activity, which is suitable for long (> 5 kb) cDNA synthesis.
One-step reverse transcription and PCR amplification are completed in the same tube without opening the tube cover between cDNA synthesis and amplification, which is helpful to reduce contamination. Since all cDNA samples obtained are used for amplification, the sensitivity is higher, with a minimum of 0.01 pg of total RNA. For successful one-step RTPCR, gene-specific primers are generally used to initiate cDNA synthesis. The two-step method, namely reverse transcription and PCR amplification is carried out in two steps. Firstly reverse transcription is carried out from an RNA template to obtain cDNA, and the obtained cDNA is subjected to one or more different PCR reactions. The two-step method can use oligo(dT) or random primers to guide the synthesis of the first strand of cDNA, and can reverse transcribe all mRNA information from a specific sample.
Products categories
WHY CHOOSE US
Since its establishment, our factory has been developing first world class products with adhering the principle
of quality first. Our products have gained excellent reputation in the industry and valuabletrusty among new and old customers..